Which Statement Is Best Supported by the Phylogenetic Tree Shown
Species W first came into existence 10 million years ago. Species W first came into existence 10 million years ago.

Phylogeny An Overview Sciencedirect Topics
A 1 and 2 B 2 and 3 C 3 and 4 D 4 and 5.
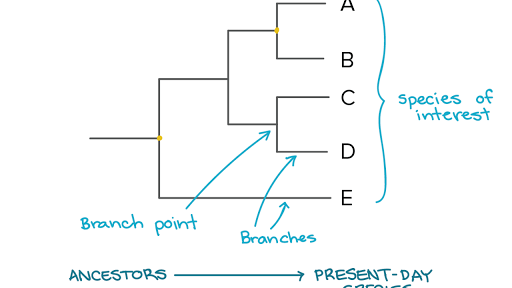
. On the contrary many evolutionary trees are so well supported and continue to be supported by newly discovered evidence that they are very likely to represent the evolutionary relationships among the organisms included accurately. Each node is called a taxonomic unit. Which statement is best supported by the phylogenetic tree shown above.
The concept of a phylogenetic tree with a single trunk representing a common ancestor with the branches representing the divergence of species from this ancestor fits well with the structure of many common trees such as the oak Figure 2012b. A total of 328 nucleotide mutation sites were found in 42 genomes among which A23403G mutation D614G amino acid change in the spike protein was the most common substitution. A phylogenetic tree of mtDNA from individuals from human populations shows that individuals from Africa are found throughout the tree while individuals from other geographic regions are generally found in only small parts of the tree.
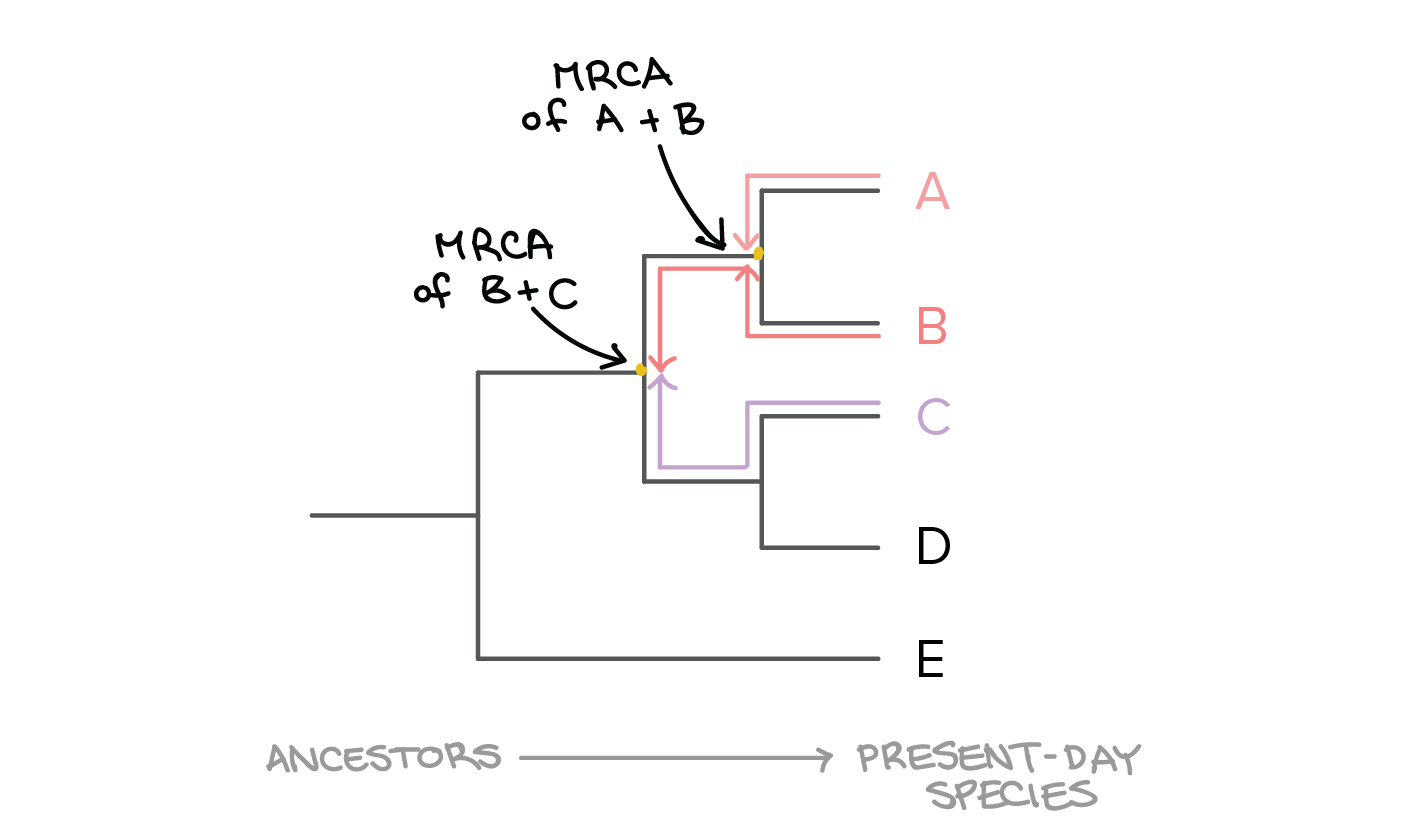
Up to 24 cash back Answer questions 5-7 using the diagram of forelimbs shown below. That node represents their most recent common ancestor. To build a cloud of trees.
C Wolves bison and horses evolved in the same habitat. AThe mastodon and the Stegodon diverged from their common ancestor 22 million years ago. Internal nodes are generally called hypothetical taxonomic units In a phylogenetic tree each node with.
We have shown two circular phylogenetic trees. Supertree method whose embryonic theory was described in the 1980s 1112 combines a set of source phylogenetic trees to produce one comprehensive phylogenetic tree reasonably that is called. If networks are tree-like one can assume that.
Which of the following claims is best supported by the information in the figure. Various methods have been developed to construct such networks using for example a multiple sequence alignment or multiple phylogenetic trees as input data. Species X Y and Z became extinct 20 million years ago.
BThe common ancestor of the African elephant and the mastodon is the Palaeomastodon. These methods are usually applied to characters but also have been used to resample taxa. Based on this phylogenetic tree which statement is true.
Ble summarizes the n-taxon statements in the RCC tree. I an 11 11 distance matrix D is constructed for the 11 species measuring differences between the row vectors of x. Phylogenetic trees show that PAO-like sequences of bacteria archaea and eukaryotes form three distinct clades with the exception of a few procaryotes that probably acquired a PAO gene through.
The phylogenetic trees of 42 SARS-CoV-2 sequences and GISAID-available SARS-CoV-2 sequences were constructed and its taxonomic status was supported. Up to 24 cash back 1. 2 shows a phylogenetic tree constructed from xThe tree-building algorithm proceeds in two main steps.
Species V is still alive today and is the oldest species. Species X Y and Z became extinct 20 million years ago. Test your understanding of most recent common ancestors with the tree shown here.
Networks show support for groups of sequences even when they are mutually incompatible and visualize edge-lengths for signal-like patterns and contradictions 3 21 22 5559. B Wolf bison and horse structures show no evidence of relationship. These two conclusions the young age of the most recent common ancestor and.
Trees based on the Pearson distance which is a statistical distance measure are less reliable than those based on Euclidean distances. Species W is still developing from a prior species. To construct the phylogenetic tree the distdna and nj commands were used supported by stats package.
CThe mammoth diverged from its most recent common ancestor with African elephants before the mastodon. Species W is still developing from a prior species. A majority rule consensus tree for groups found in 50 of the trees is used to show well-supported groups.
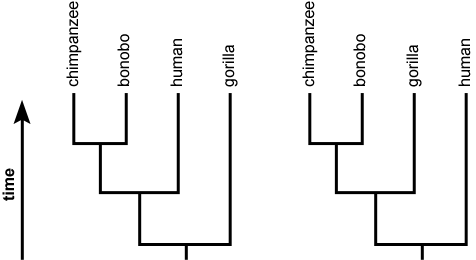
Which statement is best supported by the phylogenetic tree shown. To empirically estimate the variability. In the phylogenetic trees above numbers represent species and the same species are shown in both trees.
Species V is still alive today and is the oldest species. Our analysis of the Brassicales clade has shown that high-resolution data octanucleotides and longer sequences result in better tree topologies and higher support for branchings. However evidence from modern DNA sequence analysis and newly developed computer algorithms has caused skepticism about the.
In phylogenetics used to assess uncertainty in the proposed phylogeny. Which two species are represented as sister species in Tree 2 but are not shown as sister species in Tree 1. Coronaviruses are known to recombine frequently.
And ii D is converted into a tree by a connection algorithm that connects the closest two entries species 9 and 10 here reduces D. To find the most recent common ancestor of a set of taxa on a phylogenetic tree follow each taxons lineage back in time towards the base of the tree until all the lineages meet up. The best-supported splits by the consensus-based approach may be prohibitively large.
Rooted phylogenetic networks are used to display complex evolutionary history involving so-called reticulation events such as genetic recombination. 4 For example the idea that birds are a twig on the dinosaurs branch of the tree of life became widely accepted in the 1980s and 90s based on. Which type of animal is most closely related to a.
Which statement best represents the illustration to the right. We demonstrated the circular phylogenetic tree by using ggtree ggplot2 ape and stats packages 7 10 11. 16 Questions Show answers.
Finally phylogenetic support for a higher taxon H was measured by the mean support score of the OTUs contained within it which is the weighted sum bars_Hfracsum_kn_kHw_kHn_H where w k H is the support score for cluster k in higher taxon H n k H is the number of OTUs in cluster k and n H is the number. Phylogenetic evolutionary Tree showing the evolutionary relationships among various biological species or other entities that are believed to have a common ancestor. Evenly across preferred phylogenetic trees and most measures of support are usually given for each clade or split in a tree.
Lesser pandas are more genetically similar to giant pandas than they are to raccoons. A Wolves bison and horses are mammals.

Phylogenetic Analysis Current Biology
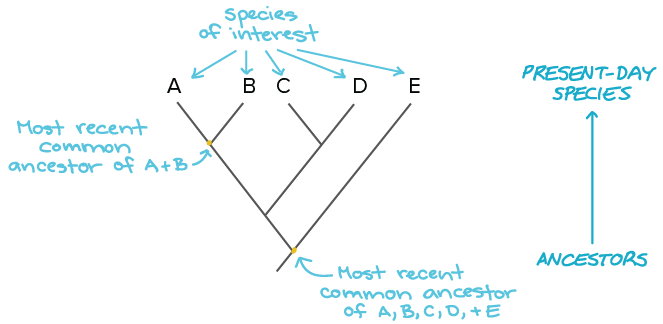
Building A Phylogenetic Tree Article Khan Academy
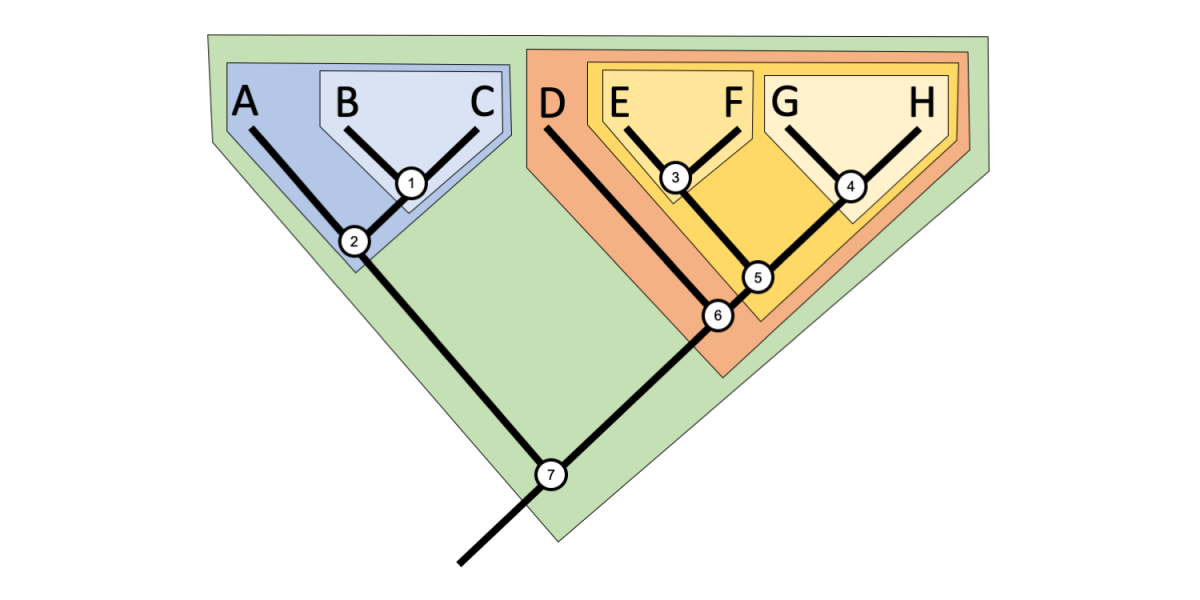
2 1 Reading Trees Digital Atlas Of Ancient Life
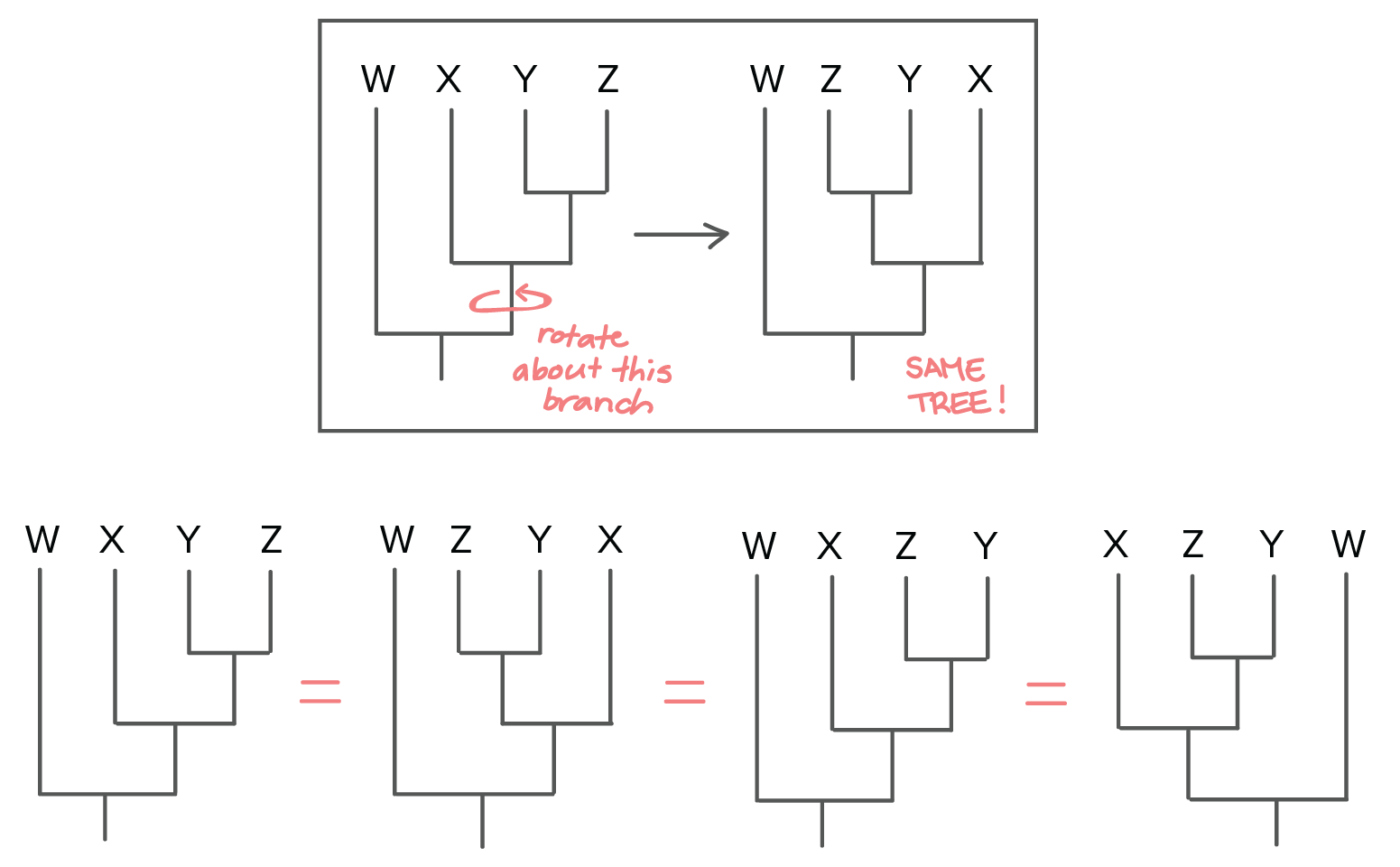
Phylogenetic Trees Evolutionary Tree Article Khan Academy
Phylogenetic Trees Evolutionary Tree Article Khan Academy
Trees As Hypotheses Understanding Evolution
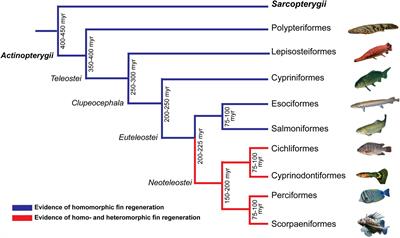
Frontiers The Fish Family Poeciliidae As A Model To Study The Evolution And Diversification Of Regenerative Capacity In Vertebrates Ecology And Evolution
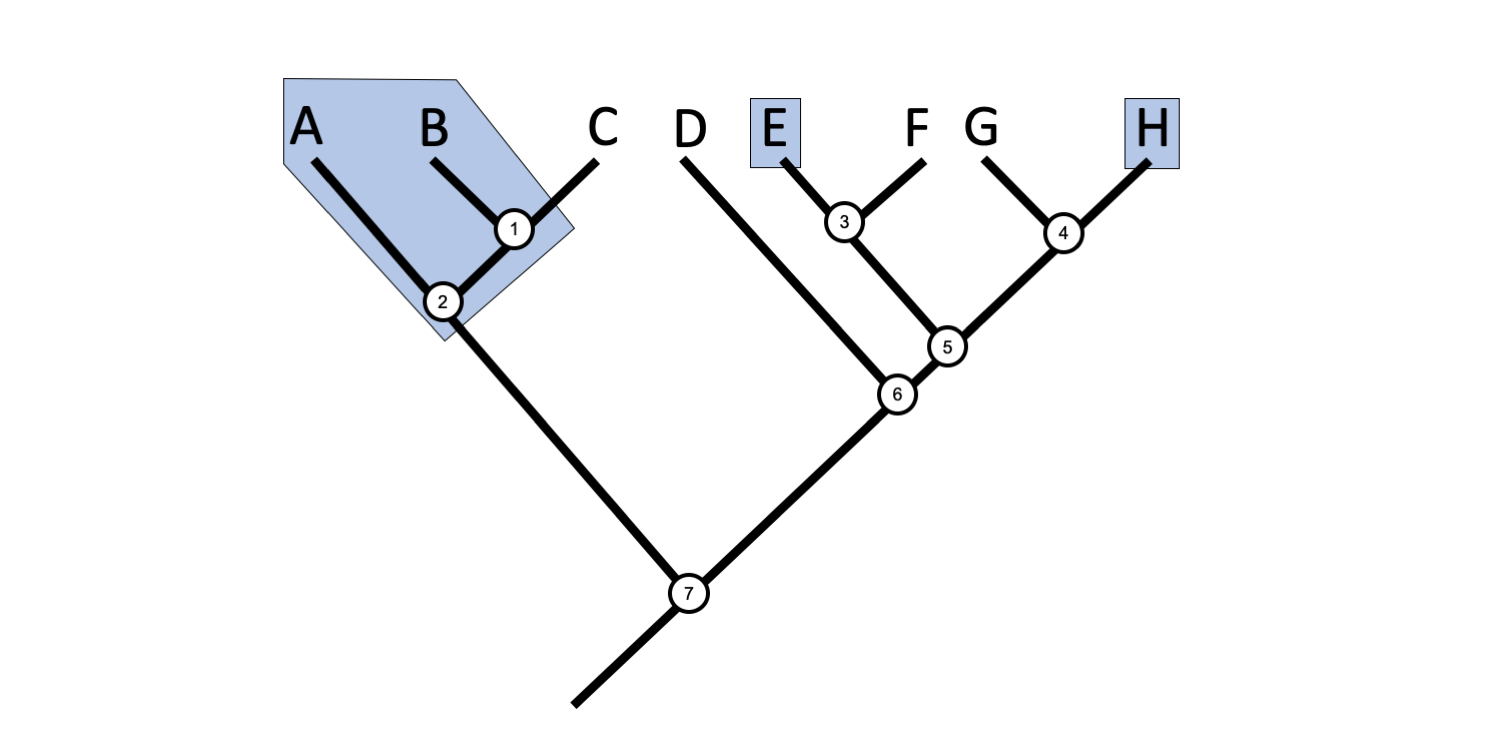
2 4 Phylogenetic Trees And Classification Digital Atlas Of Ancient Life
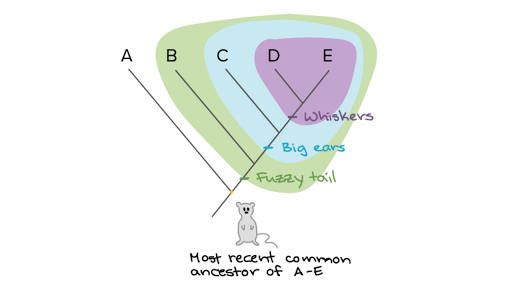
Building A Phylogenetic Tree Article Khan Academy
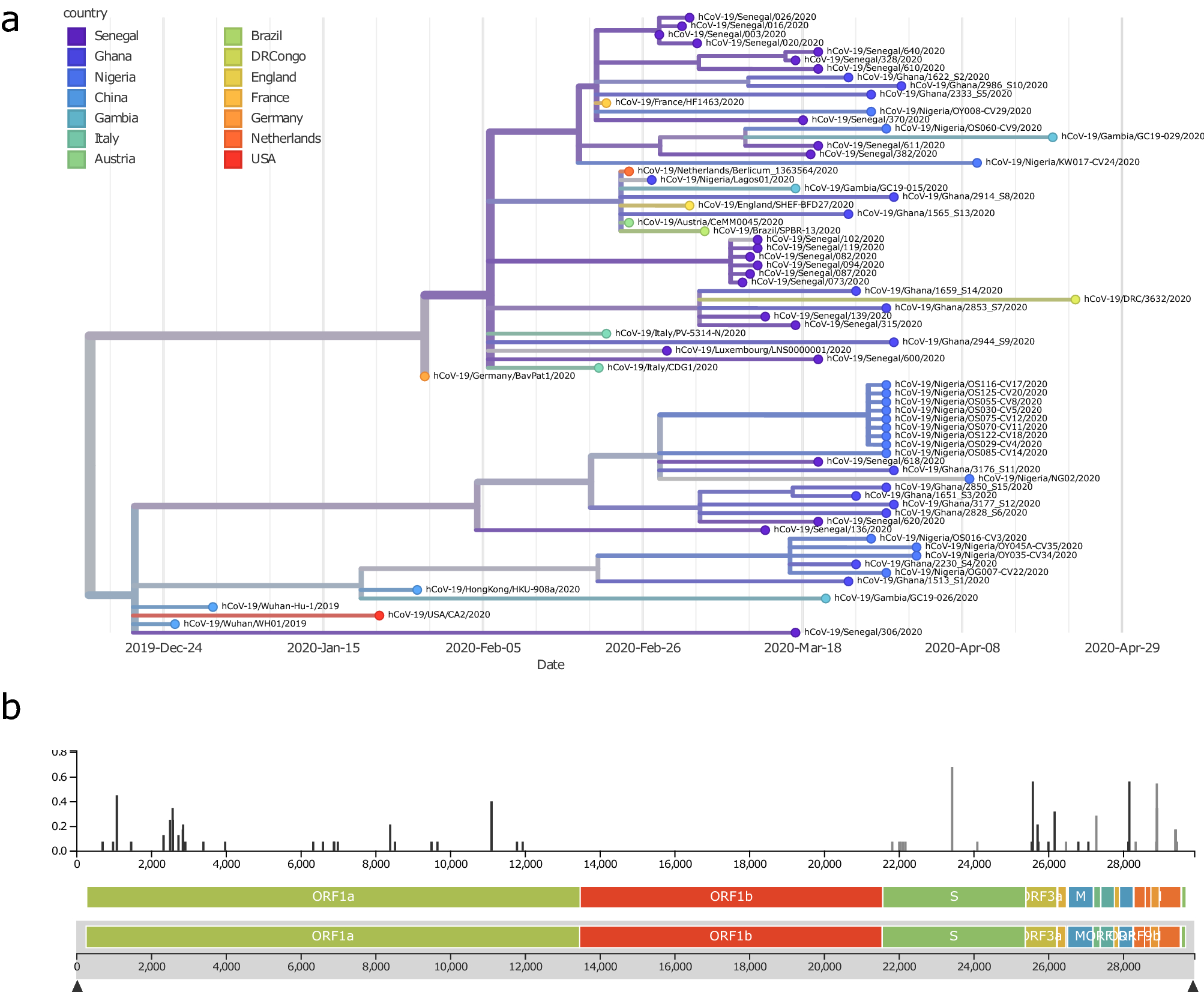
Detailed Phylogenetic Analysis Tracks Transmission Of Distinct Sars Cov 2 Variants From China And Europe To West Africa Scientific Reports
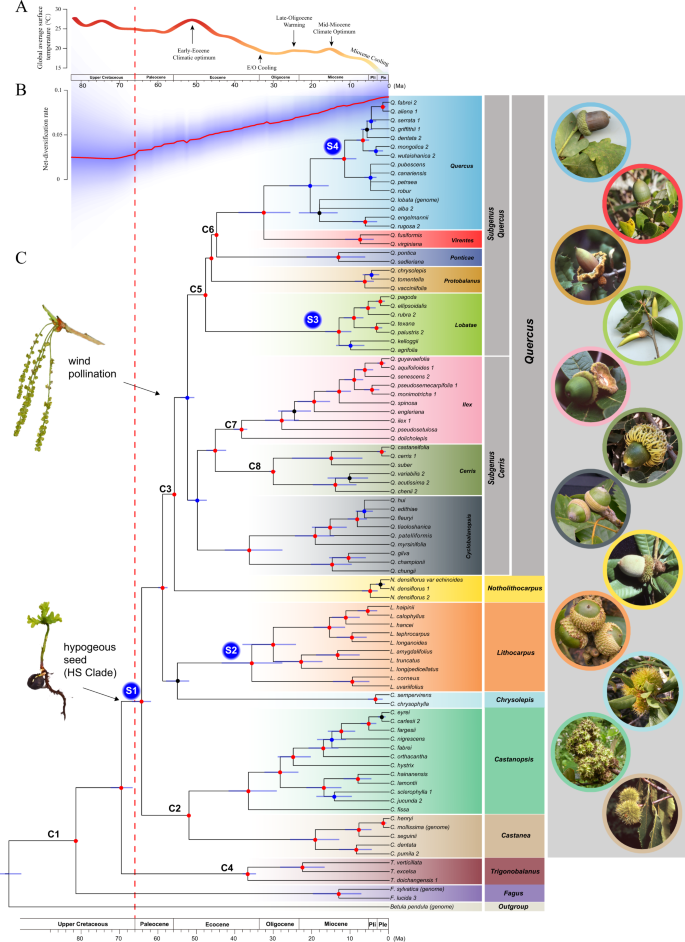
Phylogenomic Analyses Highlight Innovation And Introgression In The Continental Radiations Of Fagaceae Across The Northern Hemisphere Nature Communications

Enj Algorithm Can Construct Triple Phylogenetic Trees Molecular Therapy Nucleic Acids

Phylogenetic Trees Evolutionary Tree Article Khan Academy

Phylogenetic Tree An Overview Sciencedirect Topics
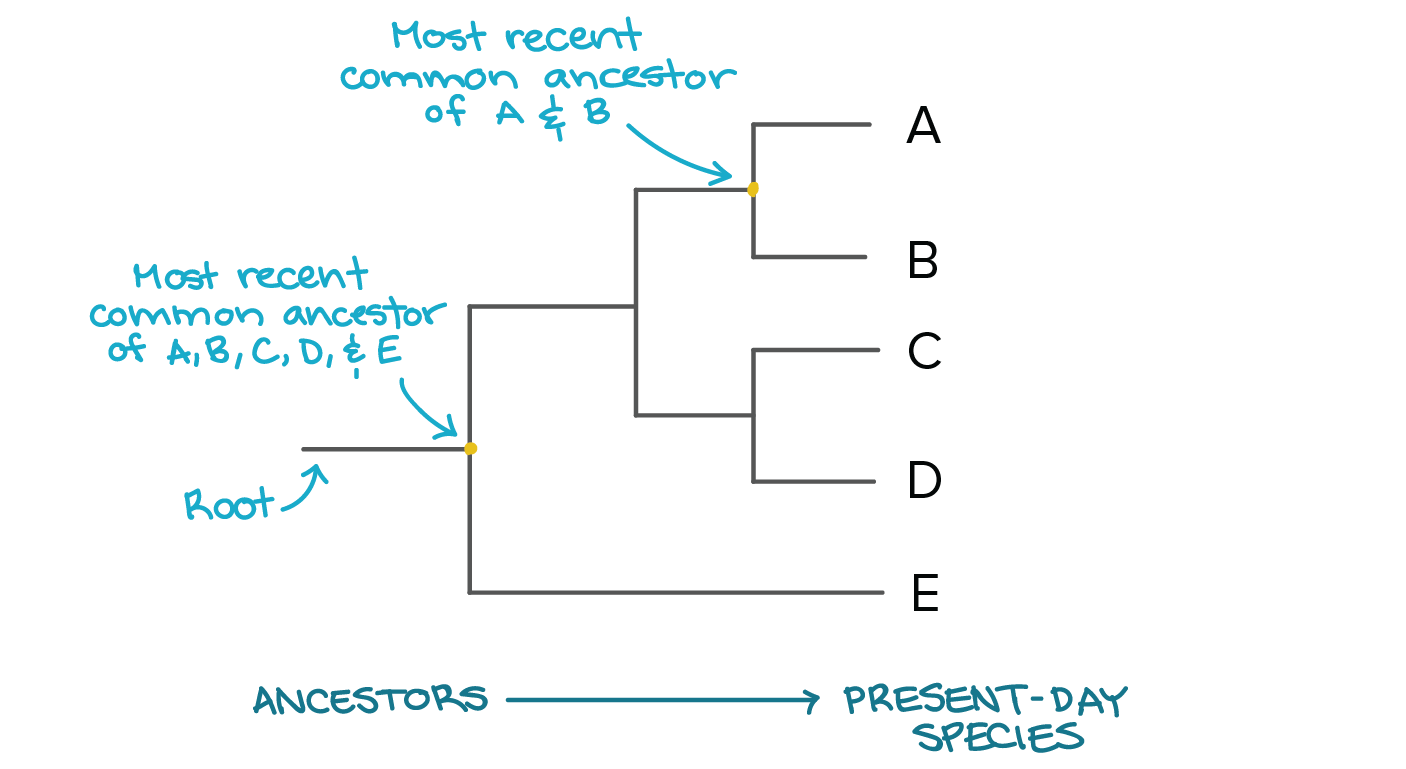
Phylogenetic Trees Evolutionary Tree Article Khan Academy
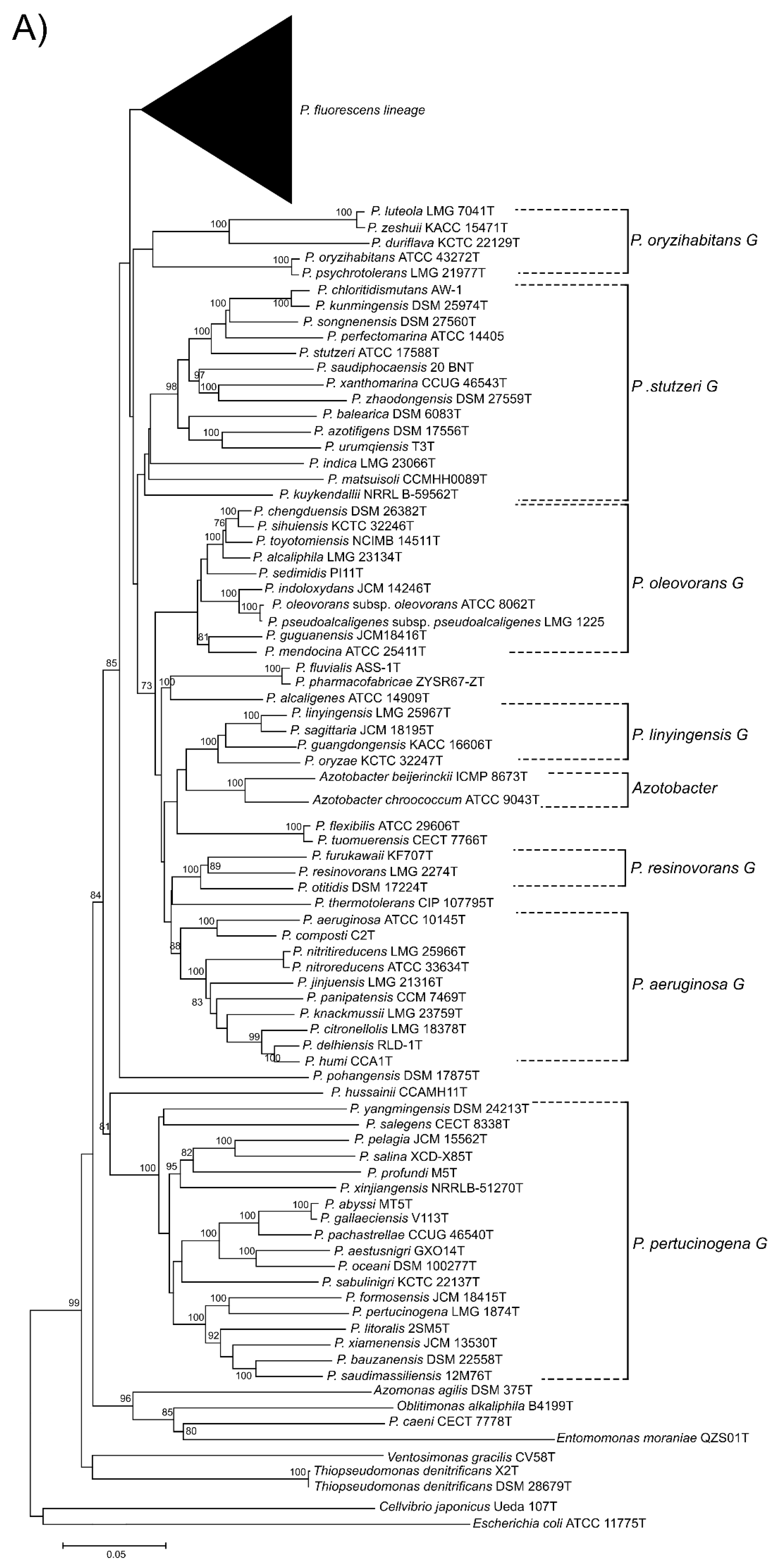
Genes Free Full Text Genomics In Bacterial Taxonomy Impact On The Genus Pseudomonas Html
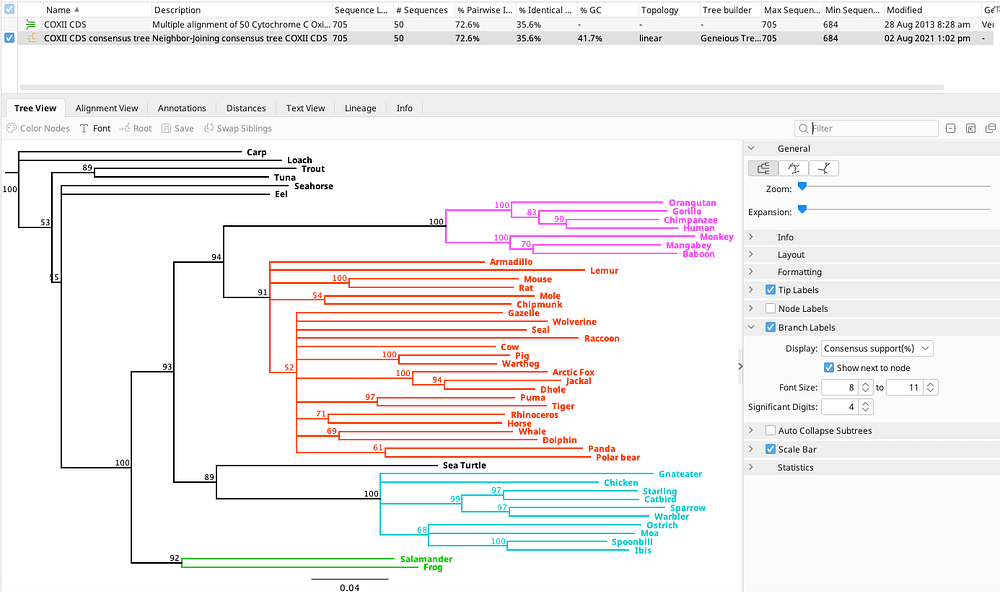
Comments
Post a Comment